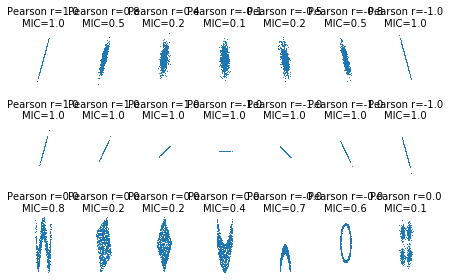
Max Information Coefficient (MIC)
Python examples on how to get MIC via minepy package can be found here: https://minepy.readthedocs.io/en/latest/python.html
import numpy as np
import matplotlib.pyplot as plt
from minepy import MINE
rs = np.random.RandomState(seed=0)
def mysubplot(x, y, numRows, numCols, plotNum,
xlim=(-4, 4), ylim=(-4, 4)):
r = np.around(np.corrcoef(x, y)[0, 1], 1)
mine = MINE(alpha=0.6, c=15, est="mic_approx")
mine.compute_score(x, y)
mic = np.around(mine.mic(), 1)
ax = plt.subplot(numRows, numCols, plotNum,
xlim=xlim, ylim=ylim)
ax.set_title('Pearson r=%.1f\nMIC=%.1f' % (r, mic),fontsize=10)
ax.set_frame_on(False)
ax.axes.get_xaxis().set_visible(False)
ax.axes.get_yaxis().set_visible(False)
ax.plot(x, y, ',')
ax.set_xticks([])
ax.set_yticks([])
return ax
def rotation(xy, t):
return np.dot(xy, [[np.cos(t), -np.sin(t)], [np.sin(t), np.cos(t)]])
def mvnormal(n=1000):
cors = [1.0, 0.8, 0.4, 0.0, -0.4, -0.8, -1.0]
for i, cor in enumerate(cors):
cov = [[1, cor],[cor, 1]]
xy = rs.multivariate_normal([0, 0], cov, n)
mysubplot(xy[:, 0], xy[:, 1], 3, 7, i+1)
def rotnormal(n=1000):
ts = [0, np.pi/12, np.pi/6, np.pi/4, np.pi/2-np.pi/6,
np.pi/2-np.pi/12, np.pi/2]
cov = [[1, 1],[1, 1]]
xy = rs.multivariate_normal([0, 0], cov, n)
for i, t in enumerate(ts):
xy_r = rotation(xy, t)
mysubplot(xy_r[:, 0], xy_r[:, 1], 3, 7, i+8)
def others(n=1000):
x = rs.uniform(-1, 1, n)
y = 4*(x**2-0.5)**2 + rs.uniform(-1, 1, n)/3
mysubplot(x, y, 3, 7, 15, (-1, 1), (-1/3, 1+1/3))
y = rs.uniform(-1, 1, n)
xy = np.concatenate((x.reshape(-1, 1), y.reshape(-1, 1)), axis=1)
xy = rotation(xy, -np.pi/8)
lim = np.sqrt(2+np.sqrt(2)) / np.sqrt(2)
mysubplot(xy[:, 0], xy[:, 1], 3, 7, 16, (-lim, lim), (-lim, lim))
xy = rotation(xy, -np.pi/8)
lim = np.sqrt(2)
mysubplot(xy[:, 0], xy[:, 1], 3, 7, 17, (-lim, lim), (-lim, lim))
y = 2*x**2 + rs.uniform(-1, 1, n)
mysubplot(x, y, 3, 7, 18, (-1, 1), (-1, 3))
y = (x**2 + rs.uniform(0, 0.5, n)) * \
np.array([-1, 1])[rs.randint(0, 1, size=n)]
mysubplot(x, y, 3, 7, 19, (-1.5, 1.5), (-1.5, 1.5))
y = np.cos(x * np.pi) + rs.uniform(0, 1/8, n)
x = np.sin(x * np.pi) + rs.uniform(0, 1/8, n)
mysubplot(x, y, 3, 7, 20, (-1.5, 1.5), (-1.5, 1.5))
xy1 = np.random.multivariate_normal([3, 3], [[1, 0], [0, 1]], int(n/4))
xy2 = np.random.multivariate_normal([-3, 3], [[1, 0], [0, 1]], int(n/4))
xy3 = np.random.multivariate_normal([-3, -3], [[1, 0], [0, 1]], int(n/4))
xy4 = np.random.multivariate_normal([3, -3], [[1, 0], [0, 1]], int(n/4))
xy = np.concatenate((xy1, xy2, xy3, xy4), axis=0)
mysubplot(xy[:, 0], xy[:, 1], 3, 7, 21, (-7, 7), (-7, 7))
plt.figure(facecolor='white')
mvnormal(n=800)
rotnormal(n=200)
others(n=800)
plt.tight_layout()
plt.show()
Part III: literature references
The following papers can also be very useful: