Accurate and fast feature selection workflow for high-dimensional omics data
citation:
Perez-Riverol Y, Kuhn M, Vizcaíno JA, Hitz M-P, Audain E (2017) Accurate and fast feature selection workflow for high-dimensional omics data. PLoS ONE 12(12): e0189875. https://doi.org/10.1371/journal.pone.0189875
key takeaways:
- Feature Selection (FS) approach is a crucial and non-trivial task because:
- it provides a deeper insight into the underlying processes that are the foundation of the data;
- it improves the performance (CPU-time and memory) of the ML step, by reducing the number of variables; and
- it produces better model results <>avoiding overfitting<>.
- A good feature subset can be defined as one that:
- contains features highly correlated with (predictive of) outcome,
- yet uncorrelated (independent) with (not predictive of) each other.
- univariate filtering
- The most-common approach - the univariate filtering - is to use a variable ranking method to filter out the least promising variables before using a multivariate method.
- However, correlation filters could prompt some loss of relevant features that are meaningless by themselves but that can be useful in combination. To overcome this effect, a set of algorithms has been proposed to combine the original variables into a new and smaller subset of features, such as Principal Component Analysis (PCA) and Linear Discriminant Analysis.
- The proposed FS workflow to perform FS in high-dimensional omics big data. The workflow combined univariate/multivariate correlation filters with wrapper feature backward elimination and it was applied to regression and classification problems.
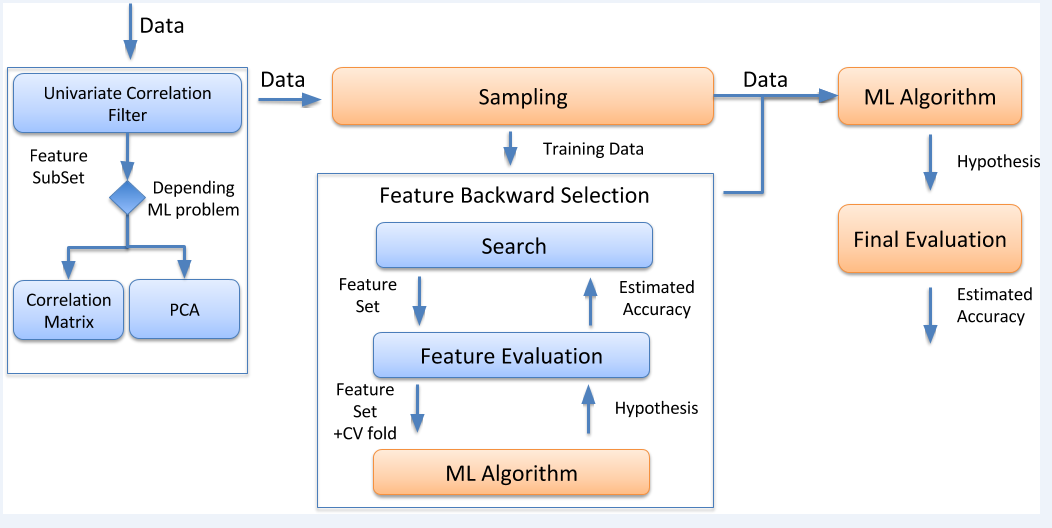
- First, a univariate correlation filter can be used before applying any wrapper approach, to determine the relation between each feature and the class or predicted variable.
- Then, a second filtering step (Correlation Matrix (CM) or PCA), can follow, in order to determine the dependencies between the different dataset features.
- Finally, backward elimination is achieved by wrapping a ML method, such as Random Forest and SVM around each example.